
The minigame tasks players with identifying highlighted cell structures from fluorescent images in exchange for ISK and Analysis Kredits that can be used to buy some shiny new Sisters of EVE items. Project Discovery can be opened from the side bar whether you’re docked or in space, making it a good way to kill some time while you’re waiting for something to happen. The task can be a bit tricky at first, but some players have already become expert classifiers with hundreds of submissions and accuracy ratings of over 90%.
In this edition of EVE Evolved, I delve into Project Discovery, link a few great community guides, and highlight some serious problems with it that have unfortunately appeared.
 What is the actual research?
What is the actual research?
You may remember the monumental news back in 2003 that the Human Genome Project had successfully sequenced all 20,000 of the protein-coding genes in a reference human genome for the first time. This was a colossal scientific achievement, but it was just the first step in a much larger scientific process. Each of those genes sequenced codes for a particular protein or piece of functional RNA, and it’s important to understand what function each of the proteins serve in a human cell. The Human Protein Atlas aims to help answer that question by figuring out exactly what parts of the cell each protein is used in.
The atlas is an important resource for scientists around the world who are working on research projects involving human genetics. Researchers working on potential treatments for genetic disorders, for example, could get clues about the mechanism behind the disorder by looking at where the affected proteins are normally expressed in a healthy cell. Figuring out where a protein is expressed in a cell can also give clues as to its function and could lead to new treatments for a variety of medical conditions.
 How to play the minigame
How to play the minigame
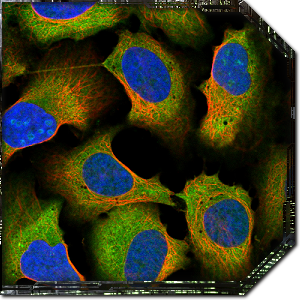
When you first launch Project Discovery, you’ll be presented with a tutorial that will walk you through some pre-classified images and explain how the interface works. You’re presented with an image of some cells with red, blue and green components, and your job is to identify which cell structures are coloured in green. The red component of the image is always cytoskeleton microtubules, the blue component is always the cell nucleus, and the green colour is the part of the cell you’re trying to identify. Each image represents a single protein that’s being investigated, and the green areas are everywhere that protein shows up.
The same protein can often be found in different parts of the cell, so the goal is to select every part of the cell that you think is being stained in green. Hovering over the various cell component options on the right hand side will show you a few example images, but you shouldn’t just classify images based on a visual match with the examples. Each cell component has an accompanying description that provides a much better explanation of exactly what you’re looking for to identify that part. For example, nucleoplasm should only be selected if it the green area overlaps completely with the blue colour, nucleoli always overlap with the holes in the blue colour, and plasma membrane is usually visible outside the red colour.
The colour options below the slide will let you toggle each individual colour on and off so that you can see whether the protein is overlapping a key feature like this. I find that it’s best to start each new image by selecting only the green colour to look for obvious features, and then toggle the red and blue on and off to look for overlaps. Remember that you can mouse over the slide to zoom in on a part of it, and click on the slide to lock that view in place so that you can have a really detailed look at the cell as you switch between the colours. If you find something that doesn’t match any of the options, you can also click the Abnormal Sample checkbox to flag the sample for review by researchers.
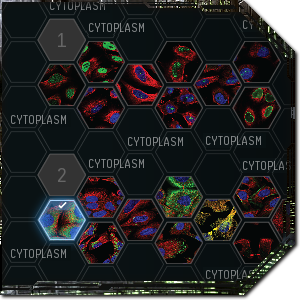 Don’t just click Cytoplasm
Don’t just click Cytoplasm
When you submit your classification, you get instant feedback showing you what percentage of players selected which options for that particular slide. This exposes some problems with the system, however, as the community often can’t make up its mind and the most popular choices are sometimes the wrong answer. The problem is that the rewards for Project Discovery are based on how quickly you can blitz through samples and whether or not your answer agrees with the community consensus, even if that consensus is wrong.
In a game like EVE where players routinely manipulate the in-game markets and have a history of exploiting game mechanics for a profit, this is potentially very dangerous. If players try to abuse the system by creating a false consensus in order to farm points, then the data retrieved from the project will not be useful. As there’s an incentive to respond as quickly as possible, most players will also select only one option even when multiple exist, so those who find multiple features are reportedly punished.
 Preventing abuse
Preventing abuse
Abuse is supposed to be prevented by the accuracy rating system, which cuts off rewards if a player’s accuracy drops below 30%. I decided to test this by creating a new character and simply clicking cytoplasm every time, and unfortunately discovered that the system is abusable. After selecting only cytoplasm for over 250 submissions, I reached rank 24 with over 10,000 Analysis Kredits and maintained a steady accuracy rating of over 55%. Every now and then, you come across a pre-classified training image that’s probably designed to catch out bots, but it’s always a very obvious and clean image and the answer is usually nucleoli.
It’s clear that even the tiny financial rewards on offer in Project Discovery can corrupt the integrity of the data being collected. We need a lot more pre-classified training images of different types mixed in to trip up people mis-identifying things, along with harsher punishments for getting them wrong and explanations like we get during the tutorial. Perhaps the immediate financial rewards should even be entirely scrapped and instead players could get rewards when they reach certain combinations of rank and accuracy rating (using only confirmed submissions that have reached a consensus). That way players who click cytoplasm all day would never get any rewards as they’d never break the 70-80% accuracy mark.
The Human Protein Atlas is an extremely worthwhile project that can make a huge difference to scientific research around the world, and gamifying it as Project Discovery was a fantastic idea. While it’s clear that the game mechanics can be abused right now, the people at HPA are really engaging with the EVE community on this and are already working to correct problems that people have brought up.
I highly recommend that every EVE player has a go at Project Discovery, but don’t do it for the tiny ISK rewards or the new Sisters of EVE swag. Do it to learn more about the different parts of the cell, to help with an incredibly important piece of scientific research, and for that slim chance of finding an abnormal sample that could lead to an interesting new discovery. Do it for science!
 EVE Online expert Brendan ‘Nyphur’ Drain has been playing EVE for over a decade and writing the regular EVE Evolved column since 2008. The column covers everything from in-depth EVE guides and news breakdowns to game design discussions and opinion pieces. If there’s a topic you’d love to see covered, drop him a comment or send mail to brendan@massivelyop.com!
EVE Online expert Brendan ‘Nyphur’ Drain has been playing EVE for over a decade and writing the regular EVE Evolved column since 2008. The column covers everything from in-depth EVE guides and news breakdowns to game design discussions and opinion pieces. If there’s a topic you’d love to see covered, drop him a comment or send mail to brendan@massivelyop.com!















